Vikrant Jandev and Marya Lieberman, UND
Blake Joachim (Outside In) and Cole Altomare-Jarczyk and Ivy Sabal (MADDS)
What’s up with the IR spectrum of BTMPS? How come the literature spectrum doesn’t fit what we are seeing in drug samples?
The literature IR spectrum of BTMPS is the free base, which is the form that is used as a plastics additive.
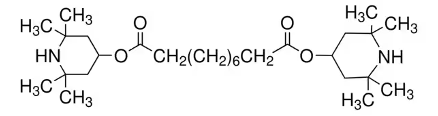
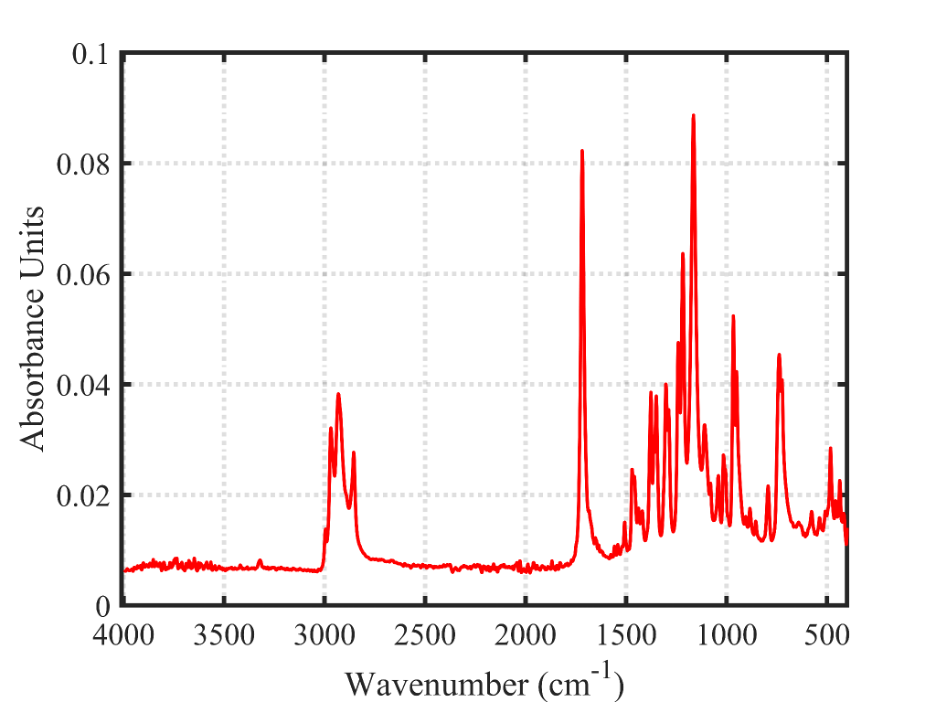
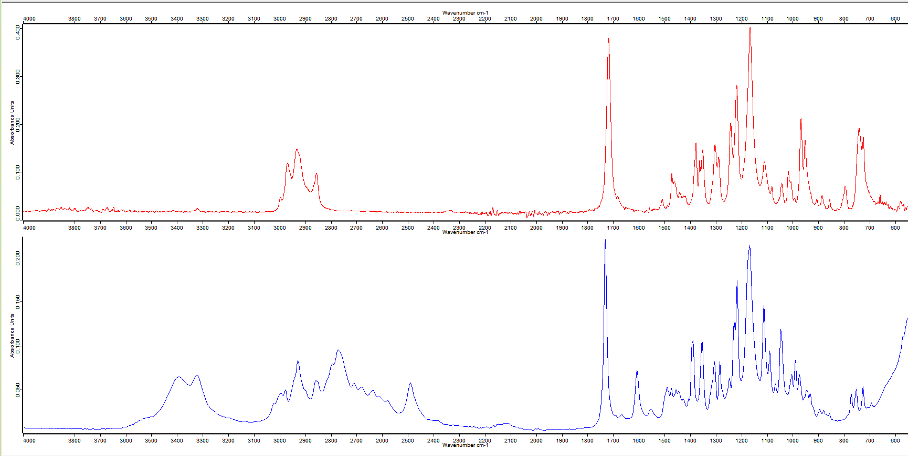
The red spectrum at the top here is the literature BTMPS spectrum, the blue spectrum at the bottom is the IR that is obtained when you subtract out the “known” components of a sample that has a lot of BTMPS, verified by GCMS.
The chemical form found in the drug supply appears to be the hydrochloride salt. Why? The hydrochloride salt of BTMPS is quite soluble in water. The free base of BTMPS is not soluble in water at all, so it would not “work” as a component of an injectable drug. (I have no clue why anyone would add it to drugs in the first place)
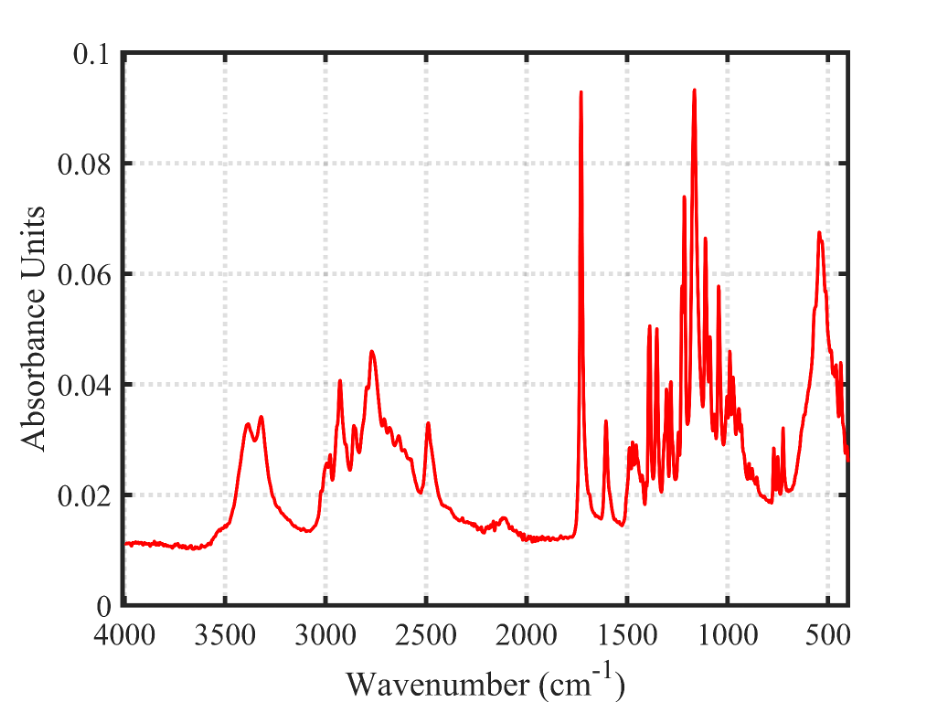
This OPUS spectrum is available for download here, in the Remedy Alliance slack channel and has been shared with MADDS, BCCSU, and Kykeon.
Synthesis: BTMPS is a “free base” meaning that the 2 nitrogen atoms in the piperidine rings are available for protonation. Addition of 2 molar equivalents of a strong acid gives the doubly protonated form of the molecule. The parent BTMPS is soluble in methanol and acetone, but insoluble in water. The hydrochloride salt is soluble in methanol and water, but not in acetone. So if you make the salt in acetone, it crashes out of solution.
1.25 g bis(2,2,6,6-tetramethyl-4-piperidyl) sebacate (BTMPS, CAS 52829-07-9, Tinuvin 770, Sigma-Aldrich #535834, 480.7 g/mol, 2.60 mmol) was dissolved in 7.5 mL acetone at room temperature giving a clear solution. 2.0 mL of 3M HCl in isopropyl alcohol was added slowly with swirling (6 mmol, 2.3 eq). The cloudy solution was refrigerated for 30 min, during which time copious white solid precipitated. The precipitate remained after warming the solution to room temperature. The solid was isolated by vacuum filtration and rinsed with 2x5 mL of RT acetone, then was dried to constant weight (1.4182 g, 98.5% assuming product is the dihydrochloride with formula C28H54N2O4Cl2, 553.65 g/mol).
Per Sigma: acetone: 19 % (w/w) at 20 °C, ethyl acetate: 24 % (w/w) at 20 °C, methanol: 38 % (w/w) at 20 °C, chloroform: 45 % (w/w) at 20 °C, methylene chloride: 56 % (w/w) at 20 °C, H2O: insoluble <6 ppm at 20 °C
While the parent BTMPS gave a sharp melting point (82.7-84.3C, compare with 82-85 °C (lit.)) the hydrochloride salt appeared to melt in several stages starting at about 118C and with solid still visible until it decomposed to a brown material at 255 C. (Catherine O’Donnell and Marya Lieberman both checked this) The infrared spectra were obtained on a Bruker Alpha II ATR-IR with 4 cm-1 resolution and 16 scans.
More resources:
1) Here is a good online tutorial about the impact of protonation on amine IR spectra: https://www.spectroscopyonline.com/view/organic-nitrogen-compounds-v-amine-salts
2) This free IR modeling site is fun to play with: Infrared spectra prediction
To use the IR modeling software, you’ll need to draw the molecule. The easiest way is to import the SMILES string. The BTMPS SMILES string is:
CC1(C)CC(CC(C)(C)N1)OC(=O)CCCCCCCCC(=O)OC2CC(C)(C)NC(C)(C)C2
Once you’ve imported the string, the structure of the molecule should pop up on the screen. The program is not powerful enough to model the entire molecule, so just erase half of the atoms (pick the left or right side, it doesn’t matter which). The IR spectrum of that half molecule will be very similar to the IR spectrum of the full molecule. Next, draw in the extra N-H bond (and add a + charge) and model the IR spectrum for the cation. Once you’ve modeled the spectrum, you can pick an IR mode and the software will then show you a movie of the corresponding bond stretches and wiggles.